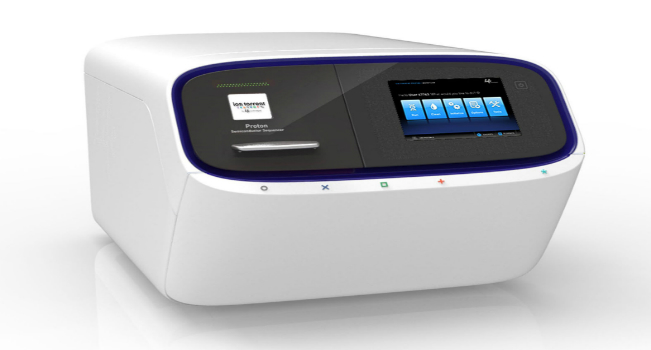
The center includes the Ion Proton as well as the Ion PGM Sequencing Platforms, with equipment for high throughput DNA extraction, DNA processing, Read More ...

The center can run whole genome sequencing, exome sequencing, as well as sequencing of gene panels. It can also run RNAseq sequencing experiments Read More ...

The sequencing experiments can be used to identify point mutations and larger structural variations against a reference genome. It can also be used to Read More ...